Abstract
Growing interest in the use of non-thermal, atmospheric pressure gas plasmas for decontamination purposes has resulted in a multiplicity of plasma-generating devices. There is currently no universally approved method of comparing the biocidal performance of such devices and in the work described here spores of the Gram positive bacterium Bacillus subtilis (ATCC 6633) are proposed as a suitable reference biological agent. In order to achieve consistency in the form in which the biological agent in question is presented to the plasma, a polycarbonate membrane loaded with a monolayer of spores is proposed. The advantages of the proposed protocol are evaluated by comparing inactivation tests in which an alternative microorganism (methicillin resistant Staphylococcus aureus—MRSA) and the widely-used sample preparation technique of directly pipetting cell suspensions onto membranes are employed. In all cases, inactivation tests with either UV irradiation or plasma exposure were more reproducible when the proposed protocol was followed.
Export citation and abstract BibTeX RIS
1. Introduction
Interest in the use of non-thermal, atmospheric pressure gas plasmas for inactivating micro-organisms has grown rapidly in recent years [1, 2, 3]. The majority of such applications are related to food [4, 5, 6] and medicine [7, 8, 9] but are by no means restricted solely to these. Over this period of time, a variety of plasma-generating devices has been described in the scientific literature [10, 11, 12] and numerous tests to inactivate a wide range of micro-organisms have been conducted. This has led to the generation of a large data resource which could in principle serve in the design of gas plasma-based disinfection processes. However, what has remained elusive is a method of rigorously comparing the biocidal performance of different plasma devices. Even determining the true biocidal potential of plasma-generating devices of apparently identical configuration is not straightforward. This can be illustrated with reference to a couple of examples. In the first of these, Xingmin et al [13] and Miao and Yun [14] using direct barrier discharges (DBDs) which were produced using similar reactor configurations, investigated the inactivation of Escherichia coli. Employing similar sample preparation methods, the former achieved a single log reduction in viability at a measured energy input of 50 J cm−2, whilst the latter achieved the same result with the much reduced energy input of 4 J cm−2. In the case of inactivation of Staphylococcus aureus, also using DBDs, Xingmin et al [13] achieved a single log reduction at an energy expenditure of just under 6 J cm−2, whereas Sun et al [15] required approximately 4.2 J cm−2, and Ma et al [16] only 1 J cm−2.
That the results from these studies are carefully obtained and valid in their own right is not contested here. Rather, the point being made is that it is difficult to compare results from different studies. It is almost certain that small differences in the operation of the devices would have had an influence on the composition of the plasma and this would have contributed to some extent to the disparity in the results of the examples cited above. Achieving stable and quantifiable plasma operation is obviously desirable, and in a recent study Pavlovich et al [17] put forward procedures designed to achieve this. This is an important aspect of experimentation with gas plasmas and one that until recently has not been given due attention. Another welcome initiative in this direction is that undertaken by the 'Plasma Sources for Biomedical Applications' working-group of the European COST action MP1101 'Biomedical Applications of Atmospheric Pressure Plasma Technology' [18], which has been working on the design and characterisation of a reference plasma source.
Notwithstanding such compositional differences, the other major factor contributing to the discrepancies in the data mentioned above arises from differences in the physical and physiological state of the bacteria presented to the plasmas. Evidence for the importance of such factors comes from a variety of previous studies. The life cycle of most micro-organisms comprises four phases; the lag phase, the exponential or log phase, the stationary phase and the death phase. Due to the expression of specific stress response genes, most bacteria possess their highest resistance to external stresses during the stationary phase [19]. That this also holds true in the case of plasma treatment was demonstrated by Yu et al [20], who reported differences in the inactivation rate constants for E. coli at mid-log, late log and stationary phases of growth.
Even when different research groups employ the same species of micro-organisms, the differences that exist between strains of that species can result in wide variations in responses to specific external stresses and this can lead to apparent discrepancies in the data obtained similar to those illustrated above. This was demonstrated, for example, in the case of the UV irradiation of a number of strains of Bacillus pumilus [21] where differences in UV resistance of up to and in excess of 300 times that of the reference strain were exhibited by environmental isolates. Similarly, Bayliak et al [22] found a wide distribution in the resistance of different strains of Saccharomyces cerevisiae to hydrogen peroxide, whilst more recently, Kossakowska et al [23] documented differences in the responses of Staphylococcus aureus strains to oxidative stress. Although the different susceptibility of strains and even mutants can be exploited to gain insights into plasma inactivation mechanisms [24], these dissimilarities indicate that for a meaningful comparison to be made, trials should be performed with not only the same species but also with the same strain.
Another important factor in determining microbial survival is the physical state in which the organisms are presented to the gas plasma. The nature of the substrate itself can have a major impact on microbial survival. This was demonstrated by Noriega et al [25] who found that Listeria innocua survived the effects of plasma treatment better when applied to the skin of chicken than on chicken flesh. This was attributed to differences in surface topography; chicken skin is characterised by the existence of feather follicles inside which the bacteria are able to avoid the lethal effects of the plasma, whereas bacteria on the surface of the chicken flesh are more exposed to the chemical species present in the gas plasma. However, even when identical substrates are employed the spatial arrangements of the cells on the substrate can also affect survival. For E. coli, Yu et al [20] observed an approximate 20-fold reduction in the inactivation rate constant as the surface cell concentration was increased from 107 to 1011.
Based on the above considerations, the objective of the present study is to propose a biological protocol for assessing the biocidal efficacy of gas plasma devices so as to enable meaningful comparisons to be made between them. Spores of the Gram positive bacterium Bacillus subtilis (ATCC 6633) are proposed as the reference biological agent and to achieve consistency in the form in which the biological agent in question is presented to the plasma, a polycarbonate membrane loaded with a monolayer of spores is proposed. The advantages of the proposed protocol are evaluated by comparing inactivation tests in which an alternative candidate organism (methicillin resistant Staphylococcus aureus—MRSA) and the widely-used sample preparation technique of pipetting cells suspensions directly onto membranes were employed. Besides plasma treatment, irradiation of bacteria-laden membranes with UV light at a wavelength of 254 nm was also conducted in order to facilitate interpretation of the results on the basis that emission from simple UV sources is intrinsically reproducible and that it remains stable over time.
2. Materials and methods
2.1. Preparation of Bacillus subtilis spore stock
The method employed to generate spores was essentially that described by Harnulv and Syngg [26] with minor variations to their procedures. Nutrient agar (Oxoid Ltd, Basingstoke, Berks) slopes of Bacillus subtilis (NCIMB 8054/ATCC 6633) were prepared from a lyophilised culture obtained from the National Collection of Industrial and Marine Bacteria (Aberdeen, UK.) using standard microbiological procedures. A loopful of cells from one such slope was used to inoculate 100 ml of Nutrient Broth (Oxoid) in a 500 ml Erlenmeyer flask which was then incubated at 37 °C for 24 h in a rotary shaker at 130 rpm. Aliquots (200 μL) of this culture were pipetted onto the surface of 140 mm diameter petri dishes containing sporulation agar formulated as described by Harnulv and Syngg [26], which were then incubated at 30 °C for 14 d. After this period of time, de-ionised water (5 mL) was carefully pipetted onto the surface of the agar and the bacterial growth was gently suspended into the water using a sterile loop. The accumulated spore suspensions from 5 petri dishes were then combined and centrifuged at 8,000 × g for 20 min using a model Z383K centrifuge (Hermle Labortechnik GmbH, Wehningen, Germany). The supernatant was then poured off and 10 mL of de-ionised water were pipetted into the centrifuge tube. The tube was then vigorously agitated to resuspend the pellet on a vortex mixer. This procedure of centrifugation and washing was repeated a further two times. The suspension was then transferred to a thin walled glass vial of 82 mm height and 27 mm dia. (Ref. 14823562, Fisher Scientific UK Ltd, Loughborough, UK). The vial was then placed in a water bath containing 20 L of water at ambient temperature (Clifton NE2-22D, Fisher Scientific UK Ltd, Loughborough, UK) and the temperature was of the bath was increased to 70 °C at a rate of 2°C min−1. Once the water bath had reached the set point (70 °C), it was maintained at this temperature for 30 min. The vial containing the spore suspension was removed from the water bath and stored at 4 °C until needed. The spore concentration was measured to be 1.5 × 1010 spores per ml.
2.2. Preparation of methicillin resistant staphylococcus aureus
A clinical isolate of MRSA (PM64) was kindly donated by Dr Julie Morrissey of the Department of Genetics, University of Leicester, UK. The culture was maintained on Luria Agar slopes and stored in a refrigerator for no more than one month before sub-culturing.
In order to prepare cultures for experiments, a loopful of cells from a slope were used to inoculate 100 ml of Luria Broth in a 500 ml Erlenmeyer flask which was incubated on a rotary shaker at 37 °C and 130 rpm for 17 h. Following this, a 1 mL aliquot of this culture was used to inoculate a fresh flask of Luria broth incubated as described above for 4 h in order to generate a mid-log phase culture (9.8 × 108 CFU ml−1). A 10 mL aliquot of this culture was transferred to a sterile centrifuge tube and centrifuged at 1750 × g for 30 min (Heraeus Labofuge 400 R, Fisher Scientific UK Ltd, Loughborough, UK). The supernatant was poured off and 10 ml of sterile Ringer's solution (Oxoid) was added to the tube. After vortexing to resuspend the pellet, the broth was centrifuged under identical conditions to those described above a further two times to yield a suspension of MRSA for deposition on membranes.
2.3. Bacterial deposition on membranes
2.3.1. Filtration—spores of B. subtilis.
The spore stock suspension generated as described above was highly concentrated, and it was found necessary to dilute it in two stages in order to obtain a homogeneous suspension of disaggregated spores. To do this 100 μl of spore stock was added to 900 μL of Ringers solution (Oxoid) in an Eppendorf tube which was then vigorously agitated using a vortex mixer. A 90 μl aliquot of this suspension was added to 10 mL of Ringers solution. This suspension was then drawn into a 10 mL syringe which was mounted onto an autoclave-sterilised Swinnex filter holder with a Lauer fitting (SX0002500, Millipore (U.K.) Limited, Watford, UK). A 25 mm diameter Nucleopore track-etched polycarbonate hydrophilic membrane with a pore size of 0.2 μm (GE Healthcare Life Sciences, Little Chalfont, UK) was loaded into the filter holder prior to autoclaving. The assembly was inserted through a rubber bung which was used to seal a heavy-walled glass conical flask of 2 L capacity and fitted with a side arm which was connected to a vacuum pump (Model N810FF-18, KNF Neuberger Inc, Trenton, USA). The arrangement is similar to that used by Bayliss et al [28] and a schematic is shown in figure 1. Filtration was conducted at 66 kPa. Following this, the membrane holder was removed from the system and disassembled to permit the membrane to be carefully removed using fine-tipped forceps and then placed in an open petri dish to dry.
Figure 1. Schematic of the arrangement used to filter-deposit cells onto membranes.
Download figure:
Standard image High-resolution image2.3.2. Filtration—MRSA.
The procedure employed with MRSA was identical to that described above for B. subtilis with the exception that 220 μL of the working suspension was pipetted into 10 ml of Ringers solution for filtration.
2.3.3. Pipetting—B. subtilis.
An aliquot (98 μL) of the working spore stock suspension was pipetted directly onto a sterile membrane placed in a petri dish. The petri dish was then placed on a hot plate (11-102-50SH Fisher Scientific UK Ltd, Loughborough, UK) at 40° C until all the liquid had evaporated.
2.3.4. Pipetting—MRSA.
The procedure outlined above was followed with the exception that 270 μL of the working suspension was pipetted onto each membrane.
2.4. Recovery of cells from membranes
To recover cells, the micro-organism laden membranes were transferred to sterile glass Universal bottles containing 10 ml Ringers solution and five 3 mm glass Ballotini beads (VWR International Ltd, Lutterworth, UK). The contents of the bottle were then vigorously agitated for 45 s using a vortex mixer. Serial dilutions in Ringers solution were carried out prior to plating 100 μL aliquots onto LB agar plates and incubating overnight at 37° C prior to counting the colonies formed. Counts were performed in triplicate.
2.5. Plasma treatment of membranes
A custom-built surface dielectric barrier discharge (DBD) plasma device was fabricated to treat the membranes (figure 2). The system is closed and operates in ambient air. The plasma source consists of a 1.6 mm thick dielectric substrate (glass-reinforced epoxy FR-4) with a powered copper plane on one side and a ground mesh-like copper electrode on the other. The membranes were kept at a distance of 9 mm from the plasma source. The plasma source was powered by an in-house built half-bridge resonant power supply that delivered a sinusoidal voltage of 5.25 kV at 20.5 kHz. The applied voltage was modulated by a square signal of 2 Hz with a duty cycle of 5%, i.e. plasma on-time of 25 ms every half second. The time-averaged power delivered to the plasma was 850 mW, as determined by the analysis of the charge versus voltage (Q–V) Lissajous diagram [27]. The bacteria-laden membranes were transferred from petri dishes to the plasma device sample holder using sterile thin-tipped forceps making sure to only contact the outer edge of the membrane where no cells were present.
Figure 2. (a) Schematic of the plasma device used to treat the spore/cell laden membranes. The membrane is clamped by an O-ring onto the base of the device. (b) View of the patterned ground electrode.
Download figure:
Standard image High-resolution imageThe maximum temperature reached during a plasma treatment was monitored using a K-type thermocouple (307P, CIE test instruments, Kolkata, IN) placed 2 mm below the plasma source. The plasma system was left to cool down to room temperature after each run. The temperature in the system increased by 7 degrees above ambient temperature after 1 h of continuous operation, but for the much shorter treatments employed for the MRSA inactivation tests, the increase in temperature was less than 2°C above the room temperature.
2.6. UV treatment of membranes
Membranes were placed inside open petri dishes for UV treatment. The petri dish was placed on a platform which rotated at 4 rpm directly underneath a 5 W mercury vapour UV source (TUV 5 W-F, Philips Belgium N.V., Brussels, BE). The rotating platform served to ensure even treatment of all areas of the membrane particularly when a stainless steel mesh (see below) was employed to reduce the UV intensity as otherwise this might have resulted in shadowing effects. The lamp was housed in a fixture which had a 2 × 9 cm aperture. The distance between the source and the base of the petri dish being 210 mm. For irradiating MRSA-laden membranes a tightly woven stainless steel mesh was placed immediately beneath the aperture to reduce the UV intensity. This was measured using a radiometer (Model UVX, UVP Ltd., Cambridge, UK). The UV intensity for irradiating B. subtilis laden membranes was 25.01 μW cm−2, whilst that for MRSA was 1.16 μW cm−2
2.7. Sample preparation for scanning electron microscope (SEM) imaging
The procedure for preparing MRSA laden membranes for SEM imaging was identical to that described by Bayliss et al [28]. Following the washing and dehydrating procedure, samples were coated with gold palladium using a SC7640 sputter-coater (Quorum Technologies Ltd, Lewes, UK) for 90 s at 25 mA. The SEM images were obtained using a TM3030 microscope (Hitachi High-Technologies Europe GmbH, Krefeld, Germany).
For B. subtilis laden membrane, the protocol was modified to avoid washing off spores during the fixation and dehydrating procedure. In particular, the first 1% gluteraldehyde and the ethanol washing stages were omitted and samples were left to dry overnight to avoid having to pour off excess solutions which had a tendency to remove 'rafts' of spores.
3. Results and discussion
3.1. Reference biological agent
Spores of the Gram positive bacterium Bacillus subtilis (ATCC 6633) are proposed as the reference biological agent. The advantages of using this particular bacterium are that it is non-pathogenic and therefore it can be widely used without high biological safety requirements, and once produced, spores stocks can be kept for many years with only a negligible reduction in viability. Although a new stock was prepared for this work in order to document precisely all the preparation steps, we have spores stocks in the lab that were prepared almost a decade ago and have over this period exhibited negligible loss of viability. This is hardly surprising given the intrinsic resistance of spores to radiation, heat and chemicals [29]. In fact, spores of the genus Bacillus (albeit B. sphaericus rather than B. subtilis), have been revived after more than 25 million years [30].
Using spores from a stock obviates the need to produce micro-organisms at a particular phase of growth for each experimental trial, which reduces variability between experiments and speeds up experimental procedures. Since spores are likely to be present in most practical applications and they are more resistant to external stresses than are vegetative bacteria, the use of spores would also add relevance to the inactivation results obtained in the proposed reference test.
Notwithstanding such considerations, it is important to ensure that the conditions under which the spores are generated is consistent as otherwise this will affect their resistance to gas plasma treatment [31]. This fact explains the exceedingly detailed procedures described under Materials and Methods for generating spore stocks and why it is necessary to adhere to them.
As a means of assessing the benefits of using B. subtilis spores as a reference biological micro-organism, inactivation tests with another bacterium have also been conducted for comparison. Methicillin resistant Staphylococcus aureus (MRSA PM64) was used in this case for its clinical relevance. Experiments involving MRSA required a Class II biological laboratory, and to minimise the sources of variability between experiments with spores of B. subtilis and those with MRSA, all the experiments reported in this work were performed in the same Class II facility. Besides the additional biological safety demands and the longer preparation time required to prepare mid-log phase bacteria for each separate experiment, MRSA is also more liable to form cell clusters. As colonies formed from cell agglomerates cannot be differentiated from those originating from single cells, clustering is a source of variability that affects plate counts. Furthermore, cell clustering also affects the consistency in which a monolayer of cells can be deposited (see section 3.2 below).
Although the plasma gas temperature can be actively controlled [32], most plasma treatments reported in the literature are conducted at room temperature without tight control over the temperature at which the cells are being treated. The resistance of vegetative cells to external stressors, however, can be quite sensitive to small temperature variations, and only a few degrees difference can drastically change the cell's response. For example, it has been documented that the antibiotic resistance of MRSA can change significantly depending on the temperature (30–37 °C) at which this is measured [33, 34]. Therefore, the use of a less temperature sensitive biological agent, such as spores of B. subtilis, provides a more robust reference test and will enable meaningful comparisons between plasma devices in different labs without the need to exercise tight control over temperature during cultivation and subsequent processing.
3.2. Deposition method and recovery rates
The physical state in which the micro-organisms are presented to the gas plasma influences their resistance to the treatment and therefore, in order to achieve meaningful comparisons between plasma devices it is important that not only the same micro-organism be used, but also that the micro-organisms are presented in a consistent and repeatable form. To accomplish this, it is proposed that membranes loaded with a monolayer of bacteria are used. The membranes provide a consistent substrate for the deposition and recovery of micro-organisms and by depositing a monolayer of micro-organisms, variability and problems associated with shadowing and stacking of bacteria are overcome [28]. Drying of membranes post deposition should be slow, i.e. at room temperature and in the absence of forced convection to avoid spores being transported across the membranes by the evaporating water film as witnessed by Bayliss et al [28].
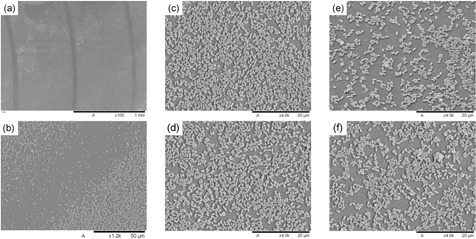
The filtration protocol described in section 2.3 was selected to deposit the number of B. subtilis spores and of MRSA bacteria required to create a monolayer on the membranes. The pipetting protocols were designed to deliver the same number of cells per unit area of membrane as for filtration. In the calculations conducted, account was made of the effective area of the membrane upon which the cells are deposited. This was because it was observed that in the case of filtration, contact of the membrane with the underlying supporting rings reduced the available area where cell could be deposited. The dimensions of these rings depend on the specific membrane holder used. These concentric rings formed on a B. subtilis spore-laden membrane are shown in figures 3(a) and (b). Most significantly, the distribution of spores across the entire membrane is fairly uniform. This is shown in the SEM images of figures 3(c)–(f) taken at four different locations across the same membrane. Two of these images where taken at regions which displayed a relatively high spore concentration (figures 3(c) and (d)) and two at regions with a relatively low spore concentration (figures 3(e) and (f)). These images clearly show that stacking of spores did not occur.
Figure 3. SEM images of a Bacillus subtilis spore-laden membrane. (a) Low magnification image showing concentric rings where spores are not deposited due to the geometric configuration of the membrane holder used during the filtration. (b) Close up view of one of the rings. (c) and (d) Close up view of two regions with high concentration of spores. (e) and (f) Close up view of two regions with low concentrations of spores.
Download figure:
Standard image High-resolution imageFigure 4 shows a comparison between the two deposition methods considered in this study, namely the proposed membrane filtering technique and the widely-used method of pipetting cell suspensions directly onto membranes. Whereas pipetting cell suspensions onto membranes results in large variations in the way cells are deposited across the same membrane (compare figures 4(a) and (b) for spores of B. subtilis and figures 4(d) and (e) for MRSA), the proposed filtering technique deposits cells uniformly across the whole membrane (with the exception of the ring regions as described above where no cells are deposited—see figure 3). The stacking and the extent of the unpopulated regions observed in the pipette-deposited membranes appear to be random as these are driven by surface tension forces acting during the drying process. It is worth mentioning that even when the density of cells in the pipetted solution is lowered, agglomeration and stacking of cells persists. This random distribution of cells affects the inactivation tests because bacteria shielded in regions where stacking occurs will be more resistant to plasma than bacteria directly exposed to the plasma in regions where few cells have been deposited. Therefore, pipetting would inevitably contribute to variability in the results of inactivation tests, preventing meaningful comparison between different plasma sources.
Figure 4. SEM images of membranes laden with (a–c) Bacillus subtilis and (d) and (e) MRSA. (a) and (b) Two regions of the same B. subtilis pipetted membrane. (d) and (e) Two regions of the same MRSA pipetted membrane. (c) B. subtilis filtered membrane. (f) MRSA filtered membrane.
Download figure:
Standard image High-resolution imageBesides sample preparation for inactivation tests, it is also imperative to assess the recovery of cells from the membranes following exposure to the biocidal agent. Figure 5 shows the percentage of cells recovered from membranes laden with B. subtilis and MRSA using the proposed filtering technique and the conventional pipetting method. The percentage of cells recovered is dependent both upon the identity of the micro-organism and the deposition method employed (figure 5), and in the case of pipetted MRSA only 9% of the deposited bacteria was recovered whereas in the case of pipetted B. subtilis 76% of the spores were retrieved. The very low MRSA recovery for pipetted samples is attributed to the tendency of MRSA bacteria to form clusters which, as explained above, in relation to plate counting will result in an underestimation of the actual number of viable cells. Although a low recovery is not a problem in itself, small variations in the low recovery rates translate into large relative errors (45% for the recovery of pipetted MRSA) and these would introduce large uncertainties in the inactivation tests.
Figure 5. Cell recovery from B. subtilis-and MRSA-laden membranes. Error bars represent standard deviation based on 4 experimental replicates.
Download figure:
Standard image High-resolution imageIt is worth noting that for both micro-organisms, recovery from filtered samples results in smaller error bars, i.e. better reproducibility, than for pipetted samples. This improved consistency in the recovery of filtered samples is attributed to the more reproducible deposition of cells across the membrane and in the reduction in the number of cell clusters present when the filtering technique is used. Although recovery of B. subtilis from filtered membranes is less effective than from pipetted membranes (probably due to some spores becoming trapped in the membrane after the pressure-driven deposition), it is evident that the smallest error bars in the recovery data (3%) were obtained for the proposed protocol of filter-deposited B. subtilis spores.
3.3. UV inactivation of B. subtilis and MRSA
Inactivation tests of B. subtilis and MRSA, both filtered and pipetted, were conducted by irradiating bacteria-laden membranes with UV light at 254 nm. The goal of these tests was to demonstrate that more repeatable results are obtained when the proposed protocol (i.e. filtered membranes with B. subtilis spores) is used.
Figure 6 shows the results of these inactivation tests. Each time point on the coloured curves was generated from three samples with counts being themselves conducted in triplicate (i.e. a total of 9 plate counts per data point on each curve). The curves depicted using the broken lines were generated from experiments conducted on separate days, and on each day a fresh stock of bacteria was prepared. The solid black line shown in the figure represents the average of all the experiments, and therefore each point is the average of 27 independent readings. Error bars represent the standard deviation from the mean. For both micro-organisms considered in this study, inactivation results determined from filtered samples are more reproducible than results obtained using pipetted samples (compare figures 6(a)–(c) for B. subtilis and figures 6(b)–(d) for MRSA).
Figure 6. Inactivation kinetics of (a) filtered B. subtilis spores, (b) filtered MRSA, (c) pipetted B. subtilis spores and (d) pipetted MRSA by 254 nm UV irradiation.
Download figure:
Standard image High-resolution imageUnsurprisingly, it is evident that micro-organisms filtered onto membranes can be inactivated more readily than those that were pipetted. For the present conditions, a further 1-log reduction is obtained for the longest treatment times (compare figures 6(a)–(c) for B. subtilis and figures 6(b)–(d) for MRSA). The lower inactivation efficacy against pipetted micro-organism is attributed to the protective shielding that occurs in regions where stacking takes place in pipette-prepared membranes (see figures 4(b) and (e)).
When comparing the inactivation curve of B. subtilis spores with that of MRSA, it is evident that the B. subtilis spores are more resistant to UV irradiation, requiring ~15 times higher UV dosage to achieve the same log reduction. It is also noted that for both, the pipetting technique and the filtering method, inactivation of B. subtilis spores were found to be more reproducible than their MRSA counterparts (compare figures 6(a) and (b) for filtered cells and figures 6(c) and (d) for pipetted cells).
In summary, of the four sample preparation methods considered in figure 6, B. subtilis spores filtered onto membranes (figure 6(a)) provides the most reproducible results.
3.4. Plasma inactivation of B. subtilis and MRSA
Inactivation tests of B. subtilis and MRSA, both filtered and pipetted, were also conducted by exposing bacteria-laden membranes to an atmospheric pressure air DBD discharge. As for the UV inactivation experiments, the goal of these tests was to demonstrate that more repeatable results are obtained when the proposed protocol is used.
Figure 7 shows the results of these inactivation tests. As before, each time point on the curves depicted using broken lines was generated from three samples with counts being themselves conducted in triplicate. The curves depicted using broken lines were generated from experiments conducted on separate days and on each day a fresh stock of bacteria was prepared. The solid black line shown in the figure represents the average of all the experiments and the error bars represent the standard deviation. For both micro-organisms considered in this study, inactivation results determined from filtered samples are more reproducible than results from pipetted samples (compare figures 7(a)–(c) for B. subtilis and figures 7(b)–(d) for MRSA). Of the four sample preparation methods considered in the plasma inactivation tests shown in figure 7, B. subtilis spores filtered onto membranes (figure 7(a)) yielded the most reproducible results.
Figure 7. Inactivation curves of (a) filtered B. subtilis spores, (b) filtered MRSA, (c) pipetted B. subtilis spores and (d) pipetted MRSA by exposure to air plasma.
Download figure:
Standard image High-resolution imageIt is interesting to note that the inactivation kinetics of filtered B. subtilis spores undergoing UV irradiation (figure 6(a)) or plasma exposure (figure 7(a)) are distinct from one another. The UV inactivation kinetics show bi-phasic characteristics whereas the plasma inactivation curve shows only a log-linear phase. This suggests that a different inactivation mechanism is at play when the cells are exposed to plasma. The tailing of the inactivation curve has been attributed to a number of factors including an intrinsic lack of precision in the enumeration of low concentration of survivors, the presence of cell aggregates, heterogeneity of the population of micro-organisms, heterogeneity of treatment, and shielding from direct exposure to the biocidal agent. It is possible that the tailing phase in the plasma-treated samples is not observed at the doses considered in this work because plasma exposure comprises diffusible gaseous agents that can exhibit lethality even against stacks of cells for which UV irradiation is ineffective.
Consideration needs to be given to the selection of a 'biological target' that is compatible with the type of plasma generating device undergoing evaluation. For a plasma jet with an effective area of a few mm2 for example, use of membranes of dimensions as specified here would be appropriate if the jet were mounted on a 2D moving stage to ensure even plasma treatment across the membrane [35]. Alternatively, the procedures described here could be applied to membranes of smaller diameter. In the same way, larger membranes could also be employed if this was warranted by the dimensions of the plasma-generating device undergoing evaluation. Whilst treatment of bacteria-laden membranes with such jets was not undertaken in this study, previous investigation has revealed that some detachment and loss of loosely adherent bacteria can occur [28] and therefore it would be essential under such circumstances to conduct appropriate control experiments in order to quantify such effects.
4. Conclusions
Spores of the Gram positive bacterium Bacillus subtilis (ATCC 6633) are proposed as a suitable reference biological agent for arriving at a valid assessment of the biocidal potential of gas plasma-generating devices The advantages of using this particular bacterium are that it is non-pathogenic, and once produced, spores stocks can be kept for many years with only a negligible reduction in viability. Using spores from a stock obviates the need to produce micro-organisms at a particular phase of growth for each experimental trial, which reduces variability between experiments and speeds up the experimental procedures. In addition, consistency in the form in which the biological agent is presented to the plasma was achieved by filtering under vacuum spore suspensions of known concentration through a polycarbonate membrane. This sample preparation technique was demonstrated to repeatedly minimise random cell stacking, a known source of variability in experimental tests.
To demonstrate the advantages of the proposed protocol, we conducted inactivation tests using another micro-organism (methicillin resistant Staphylococcus aureus—MRSA) combined with the widely used sample preparation technique of pipetting cells suspensions onto membranes. Micro-organism laden membranes were exposed to either UV light at 254 nm or atmospheric pressure air plasma. In all cases, the proposed protocol required much shorter preparation time and resulted in greater reproducibility.
Acknowledgments
This work was supported by the UK Engineering and Physical Sciences Research Council (EPSRC), Loughborough University and the University of Bologna.